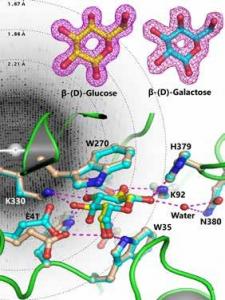
Periplasmic substrate binding proteins (SBPs) bind to a specific ligand with high affinity and mediate their transport into the cytoplasm via the cognate inner membrane ATP binding cassette (ABC) proteins. Because of very low sequence identities, understanding the structural basis of substrate recognition by SBPs has remained very challenging. A peri plasmic glucose binding protein from Pseudomonas putida CSV86 (ppGBP) is found to be highly specific towards glucose with an affinity of ~0.3 μM and has very low specificity towards galactose. No structural data was available for sugar SBPs from any Pseudomonas sp. We have determined the first high resolution crystal structures of a periplasmic glucose binding protein (ppGBP) from a Pseudomonas sp. The structures of ppGBP have been solved and refined as unliganded form (2.5 A), as well as its complexes with glucose (1.25 A) and galactose (1.8 A). Asymmetric domain closure of ppGBP has been observed upon sugar binding.The structural analysis revealed that the sugars are bound to the protein mainly by hydrogen bonds and the loss of two strong hydrogen bonds between ppGBP and galactose compared to glucose is responsible for lowering its affinity towards galactose. Interestingly, ppGBP binds only to monosaccharide but it has the structural fold of an oligosaccharide binding protein. Our structural analysis indicates that during evolution, the sugar binding pocket of ppGBP may have undergone modulation to accommodate monosaccharides only. Structural basis of sugar recognition by ppGBP. The diffraction pattern obtained from a ppGBP crystal is shown as the background. Electron density map around glucose and galactose molecules are presented on the top. Bottom portion of the figure shows structural superposition of sugar binding pockets of galactose (cyan) and glucose (yellow) bound ppGBP complexes.
Prof. Prasenjit Bhaumik
