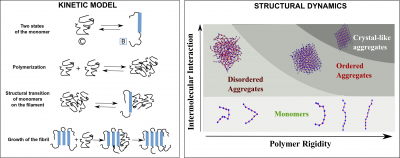
Self-assembly of bio-macromolecules into higher-order structures is a commonly observed feature in biology. Protein self-assembly in particular is associated with
a wide range of functions including cell division,cell motility and diseases such as Alzheimer’s and Parkinson’s. The forces generated during the growth of native self-assemblies such as actin and microtubules is vital for cellular function. The generation of forces is achieved by a reversible polymerisation of the constituent monomers (tubulin and actin monomers). On the other hand, aberrant self-assembly of proteins due to incorrect folding or changes in solution conditions could lead to the formation of beta-sheet rich filaments known as amyloids which could result in cell death, like in case of neurodegenerative diseases. However, similar ordered structures have also been known to perform useful functions such as reversible hormone storage, scaffold for cell growth, etc. In our group, we describe the dynamics and structural features of these molecular self-assemblies using fundamental physical principles. We provide a theoretical description of the self-assembly process using key chemical events such as polymerisation, depolymerisation, chemical modifications (GTP hydrolysis in microtubules) and structural transitions (coil to beta-sheet conversion in amyloid monomers). We identify various rate regimes under which the typical kinetic signatures in various self-assembling filaments (sigmoidal growth kinetics in amyloids, dynamic instability in microtubules, etc.) are encountered. Also, these simulations help us explore the various ‘kinetic phases’ that would result in altering the rates that govern self-assembly. For instance, under which rate regime would one observe an amyloid- like assembly as opposed to a globular aggregated structure? Similarly, what are the factors that dictate catastrophe frequencies in microtubules? We use kinetic Monte Carlo simulations and analytical theory to answer these questions. Another research focus of our group is to probe various structural features of these self-assemblies. For e.g, what are the physical factors governing the nature of the aggregate? Overall, the emphasis is on studying the kinetic and structural aspects of protein self-assemblies using computer simulations and theory. Kinetic model (left) for amyloid self-assembly which takes into account two states of the protein / peptide monomer, viz., the random coil-like state and the beta-strand state. The coil-monomers can polymerisestochastically and give rise to non-specifically bound, disordered aggregated structures. Structural transition of monomers on these aggregates stabilises the self- assembly and allow further growth to occur. Structural diversity (right) among the aggregated states as a result of varying two parameters, the inherent flexibility of the peptide / protein chain and the strength of intermolecular interactions. Various states including the monomeric state, disordered aggregates and different ordered states could be observed from these simulations.
Prof. Ranjith Padinhateeri
