Surface plasmon resonance facility

Booking Link
Registration form-External users
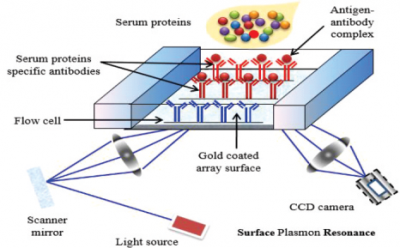
Surface plasmon resonance (SPR) is a label-free, real- time technique capable of measuring binding affinities and kinetics for bio-molecular interactions. SPR has become a key bio-sensing technology in the areas of biological research and medical sciences.
Make and Model
GE Healthcare, Biacore T200
Specifications/Features
- Can study interactions at physiologicaltemperatures. Analysis temperature range 4°C to 45°C.
- Rapid buffer scouting application for fast assay development.
- Integrated buffer degasser ensures data quality at elevated temperatures.
- Designed to support large-scale research applications
Facility in-charge
Contact Email
Location
Department of Biosciences and Bioengineering,
I.I.T. Bombay,
Powai, Mumbai - 400076
Phone Number: 022 25764757
Contact Person: Dr. Veenita Shah
Facility Management Member(s)
(w.e.f. )
Prof. Sanjeeva Srivastava
Prof. G Subrahmanyam
Prof. Samir K. Maji
Prof. Kiran Kondabagil
Prof. N S Punekar
Prof. Sarika Mehra
Prof. Ruchi Anand
Dr. M N Gandhi
