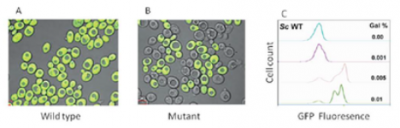
Micro-organisms have evolved an impressive array of gene regulatory mechanisms to adapt to the constantly changing environmental conditions. However, even the exquisite gene regulatory mechanisms cannot escape from the cul-de-sac of physical constraints, leading to stochastic or ‘noisy’ gene expression. Interestingly, instead of evolving mechanisms to subdue the noise and lead a detenninistic life style, micro-organisms seem to have evolved mechanisms to use this noise to their advantage. It now appears that cellular decision making is guided as much by non-genetic mechanisms arising out of noise as by genetic mechanisms.It is important to realise that deciphering the molecular basis of noise entails making sense out of the stochastic transcriptional output. To achieve this, we used a population of wild type strain of Saccharomyces cerevisiae, which responds to galactose rapidly (panel A). In contrast, a mutant strain lacking the canonical signalling gene, exhibits a significant delay in
responding to galactose. This is due to the activation of the redundant signalling pathway stochastically. Single cell analysis indicated that the inoculum of mutant cells contains only 3 rapid responders to galactose out of 1000. Because of this, as the population of mutant cells grows in galactose, responders try to overpopulate non-responders (panel B) while wild type population consists of only responders (panel A in figure).Further experiments aimed at understanding this unusual phenomenon indicated that stochastic gene expression is in turn a detenninistic process controlled by the intricate features of a genetic switch. By changing the genetic background of the mutant strain, the number of rapid inducers increased from 3 to 50 out of 1000 cells. Using FACS analysis, we recently demonstrated that even the wild type strain
exhibits non-genetic heterogeneity (panel A). Thus, our ability to track and tweak single cell behaviour in a population of genetically identical cells allows us to predict the behaviour of a population. From these studies, it appears that non-genetic mechanisms help microbial cells to transit smoothly from one nutrient condition to another without having to go through the process of adaptation. Thus, deciphering the contribution of gene expression noise to single cell behaviour is the key to evaluating the overall population behaviour. These observations have ramification in understanding many facets of applied and basic biology.
Prof. P J Bhat
