DNA is a very long ribbon-like polymer that contains the genetic code. Even though different cell types in our body (skin cells, muscle cells, brain cells, etc.) have exactly the same DNA, these cells function very differently. How is this achieved is not well understood. We now know that the fate of a cell is not just decided by the sequence of the DNA but also by the ‘state’ of its chromatin. Chromatin is a 3-dimensional active assembly of DNA bound by many proteins (a set of bio-polymer molecules). Chromatin can be assembled in multiple ways. As our skin cells have a different chromosome organisation compared to our brain cells this eventually results in different function. In other words, regulating its organisation, different parts of the DNA (genes) are made inaccessible (off) and accessible (on) leading to different types of cell and functions.In our lab, we do theoretical / computational studies to investigate different types of plausible chromatin organisations and their implications.
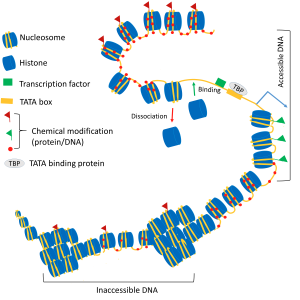
At the primary level, DNA polymer is wrapped around many ball-like proteins (known as nucleosomes) to assemble chromatin in the form a ‘string of beads’. The precise positioning of nucleosomes in space and time is crucial as it can regulate the accessibility of DNA-binding proteins to the genetic code. Using computer simulations and theoretical tools from physics and chemistry, we investigate the following questions:
■Given the experimentally known positioning of these nucleosome beads, can we derive a set of rules obeyed by the self-assembling nucleosome beads?
■How do the positioning and dynamics of these nucleosomes affect the switching ON and OFF of genes? In other words, how do nucleosomes take part in the ‘decision making’ process in a cell?
■How do nucleosomes affect the 3-dimensional organisation of the genome?
■Can we build a computer program that can predict the ‘state’ of the chromatin, under a given set of conditions, and hence help in predicting the fate of a cell?
Prof. Ranjith Padinhateeri
